Hemiselmis
John M. Archibald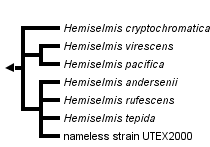

This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxCharacteristics
- Types of biliprotein: PC630, PC612, PC612 or PE555 (with subtype PE545/555)
- No dimorphism known, but differences in cell sizes have been observed in some cultures
- Light microscopy: blue to blue-green or red cryptophytes, small, often reniform cells (< 10 µm in cell length) with laterally inserting flagella, one plastid, predominantly marine (one freshwater species described: Hemiselmis amylosa; Clay and Kugrens 1999)
- Ultrastructure: hexagonal plates on both sides of the periplast, short gullet, thylakoids traverse the pyrenoid matrix, nucleomorph free in periplastidial space (Santore 1984)
Discussion of Phylogenetic Relationships
The type strain of Hemiselmis rufescens, established and examined by Parke, is still available from the Plymouth Algal Culture Collection (UK), and thus was genetically characterized in the revision of the genus Hemiselmis (Lane and Archibald 2008). Hemiselmis appears to represent a consistent genus in molecular phylogenies without any evidence for dimorphism thus far (Figure 1; Lane and Archibald 2008). At least four different biliprotein types, three blue phycocyanins and one red phycoerythrin, can be found (Lane and Archibald 2008; Hoef-Emden 2008). The reddish Hemiselmis rufescens and Hemiselmis andersenii evolved from blue-green Hemiselmis species (Lane and Archibald 2008). The problems inherent in the morphospecies concept also apply to Hemiselmis: molecular phylogenetic trees show a higher genetic variation than the morphological characters do.

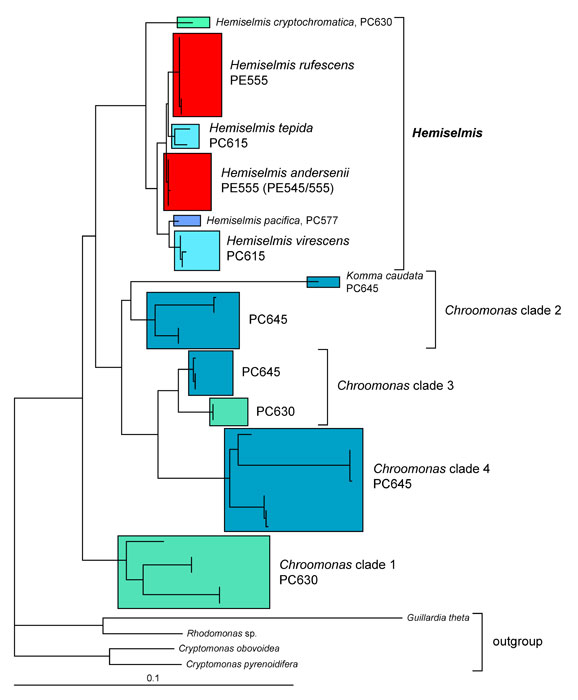
Figure 1. Molecular phylogeny of the genera Chroomonas, Hemiselmis and Komma. Maximum likelihood tree inferred from nucleomorph SSU rDNA sequences. PC, phycocyanin; PE, phycoerythrin; scale bar = substitutions per site. © 2008 Kerstin Hoef-Emden
Nucleomorph Genome
Preliminary analyses of the nucleomorph genomes of several members of the genus Hemiselmis have been performed (Lane and Archibald 2006, 2008). As in the model cryptophyte Guillardia theta (Douglas et al. 2001), all examined Hemiselmis species possess three nucleomorph chromosomes. Estimated genome sizes for the group range from ~560 kb in H. tepida to ~600 kb in H. rufescens. Unlike G. theta, which possesses complete and near identical ribosomal DNA operons on both ends of each nucleomorph chromosome (Douglas et al. 2001; Archibald 2007), Hemiselmis species are characterized by the absence of 18S and 28S loci on chromosome II (only the 5S locus is present on all 6 chromosome ends in examined species). Nucleomorph karyotype similarities within and between Hemiselmis species correlate with the molecular phylogenetic analyses performed thus far (Lane and Archibald 2008).
The nucleomorph genome of Hemiselmis andersenii CCMP644 has been completely sequenced (Lane et al. 2007). It is 571,872 bp in size and possesses 525 genes (472 protein genes and 53 non-mRNA genes). It encodes the identical set of 30 plastid-targeted proteins as Guillardia theta and also possesses a high gene density (1.09 kb/gene). The most striking feature of the H. andersenii genome is the complete absence of spliceosomal introns and genes for splicing RNAs and proteins (Lane et al. 2007). The G. theta nucleomorph genome possesses 17 small introns (42-52 bp) and genes for spliceosomal components (Douglas et al. 2001).
References
Archibald JM (2007) Nucleomorph genomes: structure, function, origin and evolution. BioEssays 29: 392-402
Clay B, Kugrens P (1999). Characterization of Hemiselmis amylosa sp. nov. and phylogenetic placement of the blue-green cryptomonads H. amylosa and Falcomonas daucoides. Protist 150: 297-310
Douglas SE, Zauner S, Fraunholz M, Beaton M, Penny S, Deng LT, Wu X, Reith M, Cavalier-Smith T, Maier UG (2001) The highly reduced genome of an enslaved algal nucleus. Nature 410: 1090-1096
Hoef-Emden K (2008) Molecular phylogeny of phycocyanin-containing cryptophytes: Evolution of biliproteins and geographical distribution. J. Phycol. 44: 985-993
Lane CE, Archibald JM (2006) Novel nucleomorph genome architecture in the cryptomonad genus Hemiselmis. J. Eukaryot. Microbiol. 44: 339-350
Lane CE, Archibald JM (2008) New marine members of the genus Hemiselmis (Cryptomonadales, Cryptophyceae). J. Phycol. 44: 339-350
Lane CE, van den Heuvel K, Kozera C, Curtis BA, Parsons B, Bowman S, Archibald JM (2007) Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc. Natl. Acad. Sci. USA 104: 19908-19913
Parke M (1949) Studies on marine flagellates. J. Mar. Biol. Ass. UK 28: 255-285
Santore UJ (1984) Some aspects of taxonomy in the Cryptophyceae. New Phytol. 98: 627-646
About This Page
This page is being developed as part of the Tree of Life Web Project Protist Diversity Workshop, co-sponsored by the Canadian Institute for Advanced Research (CIFAR) program in Integrated Microbial Biodiversity and the Tula Foundation.
John M. Archibald

Dalhousie University, Halifax, Nova Scotia, Canada
Correspondence regarding this page should be directed to John M. Archibald at
Page copyright © 2010 John M. Archibald
All Rights Reserved.
- First online 14 September 2008
- Content changed 14 September 2008
Citing this page:
Archibald, John M. 2008. Hemiselmis . Version 14 September 2008 (under construction). http://tolweb.org/Hemiselmis/97239/2008.09.14 in The Tree of Life Web Project, http://tolweb.org/





 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site