Papilionoideae
Martin F. Wojciechowski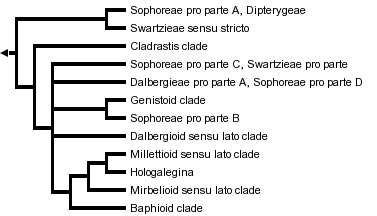

This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxIntroduction
Papilionoideae (the "papilionoids"), the largest of the three subfamilies of Fabaceae with 476 genera and 13,860 species, is also the most diverse and widely distributed, and includes most of the familiar domesticated food and forage crops and model genetic/genomic species (Gepts et al., 2005).
Characteristics
Papilionoideae is characterized by pentamerous, zygomorphic papilionoid (pea-like) flowers that have differentiated an adaxial petal (banner or standard) that is external to the others in bud, and abaxial and lateral pairs of petals (keel and wings, respectively), with the keel petals fused to enclose the stamens and gynoecium. Although this floral architecture is ubiquitous in the subfamily, exceptions do occur, with nearly actinomorphic flowers (e.g., Cadia in tribe Sophoreae; Inocarpus in tribe Dalbergieae) and flowers lacking 1 or more of the five petals known from several groups (e.g., tribe Amorpheae).
Unlike the other subfamilies, many papilionoids have the ability to synthesize quinolizidine alkaloids, isoflavones, and non-protein amino acids such as canavanine, while bipinnate leaves, complex leaf glands and compound pollen grains are lacking (Polhill, 1994).
Discussion of Phylogenetic Relationships
Although the majority of tribes are still defined in a more traditional sense (e.g. Polhill, 1994) the results of phylogenetic studies from the last 10 years necessitate quite radical shifts in inter- and intra-tribal relationships within the subfamily, and these insights have profound implications with regard to papilionoid evolution.
Papilionoideae has been consistently resolved (and strongly supported) as monophyletic in analyses of molecular data (e.g., Doyle et al., 1997, 2000; Pennington et al., 2001; Wojciechowski et al., 2004), but on morphological criteria the uncertainty regarding the position of papilionoid tribes such as Swartzieae, long considered 'transitional' between Caesalpinioideae and Papilionoideae, has clouded this issue. Rather than being resolved as the sister group to an isolated caesalpinioid lineage(s), as are the mimosoids, papilionoids are sister to the large clade comprised of Caesalpinieae sens. lat., Cassieae sens. strict., and Mimosoideae in the legume phylogeny (Wojciechowski et al., 2004). Although papilionoids have been considered to contain most of the "derived" groups within the family (e.g., predominantly herbaceous groups), fossil evidence and divergence time estimates clearly indicate the oldest crown clades within papilionoids are comparable in age (39 to 59 Ma) to the oldest caesalpinioid diversifications (Lavin et al., 2005).
Based on results from such studies papilionoids can be broadly divided into the following main subclades and groups: Swartzieae sens. str., the Cladrastis clade, the Genistoid clade, the Dalbergioid sens. lat. clade, Mirbelioid sens. lat. clade, Millettioid sens. lat. clade, Hologalegina, and several weakly supported and/or partially resolved groups such as the "basal papilionoid" groups (including members of tribes Swartzieae, Sophoreae, Dalbergieae and Dipterygeae), and the Baphioid clade.
References
Doyle, J.J., J.L. Doyle, J.A. Ballenger, E.E. Dickson, T. Kajita, and H. Ohashi. 1997. A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Amer. J. Bot. 84: 541-554.
Doyle, J.J., J.A. Chappill, C.D. Bailey, and T. Kajita. 2000. Towards a comprehensive phylogeny of legumes: evidence from rbcL sequences and non-molecular data. Pages 1-20 in Advances in legume systematics, part 9, (P. S. Herendeen and A. Bruneau, eds.). Royal Botanic Gardens, Kew, UK.
Gepts, P., W.D. Beavis, E.C. Brummer, R.C. Shoemaker, H.T. Stalker, N.F. Weeden, and N.D. Young. 2005. Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol. 137: 1228 ? 1235.
Lavin, M., P.S. Herendeen, and M.F. Wojciechowski. 2005. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst. Biol. 54: 530-549.
Pennington, R.T., M. Lavin, H. Ireland, B.B. Klitgaard, and J. Preston. 2001. Phylogenetic relationships of basal papilionoid legumes based upon sequences of the chloroplast trnL intron. Syst. Bot. 26: 537-566.
Polhill, R.M. 1994. Classification of the Leguminosae. Pages xxxv?xlviii in Phytochemical dictionary of the Leguminosae (F. A. Bisby, J. Buckingham, and J. B. Harborne, eds.). Chapman and Hall, New York, NY.
Wojciechowski, M. F., M. Lavin, and M. J. Sanderson. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. American Journal of Botany 91: 1846-1862.
Title Illustrations
| Scientific Name | Vigna speciosa (Kunth) Verdc. |
|---|---|
| Location | Mesa, Arizona |
| Comments | Commonly called Snail Vines. |
| Specimen Condition | Live Specimen |
| Copyright | © |
| Scientific Name | Erythrina amazonica Krukoff |
|---|---|
| Location | Ecuador |
| Specimen Condition | Live Specimen |
| Copyright | © Robin Foster |
| Scientific Name | Calia secundiflora (Ortega) Yakovlev (= Sophora secundiflora) |
|---|---|
| Location | Arizona State University campus |
| Specimen Condition | Live Specimen |
| Identified By | M F Wojciechowski |
| Copyright |
© Martin F. Wojciechowski

|
About This Page
Martin F. Wojciechowski

Arizona State University, Tempe, Arizona, USA
Correspondence regarding this page should be directed to Martin F. Wojciechowski at
Page copyright © 2006 Martin F. Wojciechowski
- First online 14 June 2006
- Content changed 14 June 2006
Citing this page:
Wojciechowski, Martin F. 2006. Papilionoideae. Version 14 June 2006. http://tolweb.org/Papilionoideae/60240/2006.06.14 in The Tree of Life Web Project, http://tolweb.org










 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site