Eukaryotes
Eukaryota, Organisms with nucleated cells
David J. Patterson and Mitchell L. Sogin

This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxTo date, about 60 lineages of eukaryotes have been identified (Patterson 1999). The relationships among these are not clear. In the tree above, we identify some of the more familiar lineages, including those groups which contain the most diverse multicellular eukaryotes (plants, animals and fungi). The remaining lineages are grouped together on a separate page for practical purposes.
On an accessory page, we provide an overview of the major lineages of eukaryotes.
Introduction
The eukaryotes include ourselves, other animals, plants and fungi and a rich variety of micro-organisms (protists). The protists include parasites that are so successful as to compromise the economies of entire countries. Eukaryotes visibly influence the nature of our world. Our concepts of beauty, logic and spirit are arguably derived from our observations of the eukaryotes.
The Eukaryota - along with the Archaea and Eubacteria - make up the three major branches of living organisms (viruses excepted). Eukaryotes are usually distinguished from other forms of life by the presence of nuclei and the presence of a cytoskeleton. The nuclei contain genetic information which is organized into discrete chromosomes and contained within a membrane-bounded compartment. The word 'eukaryote' means 'true nuclei'. The cytoskeleton is a complex array of proteins which provides the structural framework for the eukaryotic cell and its components, inclusive of the nucleus. The cytoskeleton has been widely exploited in the evolutionary diversification of eukaryotes. The cytoskeleton and organelles are discussed in more detail in the 'Characteristics' section below.
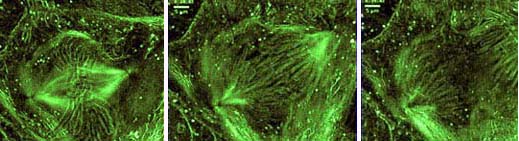
Three stages of movement of chromosomes during division of a nucleus in a newt lung epithelial cell.
Photograph copyright ? 1998 Rudolf Oldenbourg.
Eukaryotes probably emerged from prokaryotic ancestry about 1.6 - 2.1 billion years ago (Knoll, 1992). The evolutionary diversification of eukaryotes has involved invention of organelles, and their modification. Now, about 60 types of eukaryotes can be distinguished on the basis of their cellular organization (Patterson, 1999). One of these lines (the opisthokonts) includes the million or so species of animals and the fungi, another (the Viridaeplantae) is formed of the green algae and the land plants. The majority of the lines of eukaryotes are traditionally classified in a paraphyletic assemblage of mostly unicellular organisms referred to as the protists.
Characteristics
What makes a eukaryote a eukaryote? The eukaryotes are distinguished from prokaryotes by the structural complexity of the cells - characterized by having many functions segregated into semi-autonomous regions of the cells (organelles), and by the cytoskeleton.
The most evident organelle in most cells is the nucleus, and it is from the presence of this organelle that the eukaryotes get their name. Most cells have a single nucleus, some have more (some have thousands) and others like red blood cells of ourselves have none - but they can be shown to derive from cells with nuclei. Nuclei contain most of the genetic material of a cell - with other elements of the genome located in mitchondria and plastids (if those organelles are also present). The nucleus is bounded by a membranous envelope. The nuclear envelope is part of the endomembrane system that extends to include the endoplasmic reticulum, dictyosomes (Golgi apparatus) and the cell or plasma membrane that encloses the cell. The envelope is perforated by nuclear pores which allow compounds to pass between the nucleus and the surrounding cytoplasm. Some protists have more than one kind of nucleus - using one to retain a copy of the genome for purposes of reproduction, and another in which some genes have been greatly amplified, to regulate activities. Within the nucleus, the genes are located on a number of chromosomes. The total amount of DNA in a nucleus measuring less than one hundredth of a millimetre across may stretch to over a metre. When not in use this is kept within a nucleus measuring only a few microns across by being bundled up in superhelical arrays.
The cytoskeleton is comprised of a rich array of proteins. The major ones are tubulin (which forms microtubules) and actin (forming microfilaments) and a myriad of interacting proteins which effect movement or create the skeletal architecture of cells. The cytoskeleton provides shape for the cell and support for membranous organelles. It also provides anchorage for motility proteins which transport materials within the cell and cause deformations which bring about the movements of the entire cell - or organism.
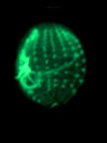
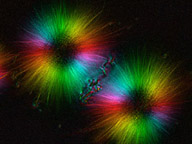
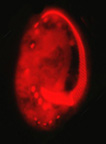
Examples of cytoskeletal architecture: metaphase chromosomes (left), kinetids (center), cytoskeleton of Euplotes (right).
Left and right photographs copyright ? 2000 David J. Patterson. Center photograph copyright ? 1996 Rudolf Oldenbourg.
Many metabolic functions are carried out within membrane bound organelles. The organelles include the endoplasmic reticulum, dictyosomes (= Golgi apparatus), lysosomes, and peroxisomes. Other membrane bound organelles that are not always present include chloroplasts (in plants, algae and organisms which have developed symbiotic associations with plastids, algae or plants) and mitochondria or hydrogenosomes. Protists (mostly microbial eukaryotes) have membrane-bounded organelles not found in most other eukaryotes - such as contractile vacuoles and extrusomes. Non-membrane-bound organelles include cytoskeletal elements (such as microtubular constructs made up of tubulin, or filamentous structures often incorporating actin), contractile systems (actin-myosin assemblages, spasmin/centrin) or other motility devices (mitotic spindle, myonemes, cilia, flagella).
The Protista
The protists are a paraphyletic group consisting of those eukaryotes which are not animals, true fungi or green plants. Using ultrastructural characteristics, we can identify about 60 types of protists, and the relationships among these lineages are not clear.
There are estimated to be about 200,000 named species of protists. Some of the groups of protists contain only one or a few genera or species, but others encompass an immense diversity of organizational types (including multicellularity) which eclipses that found in some of the more familiar animals or plants. A particularly good example of this is the territory referred to as the stramenopiles. This group embraces a quantity of photosynthetic activitiy second only to the land plants, and it includes fungal like organisms (Oomycetes), parasitic protozoa (opalines and Blastocystis), free-living protozoa (some heliozoa and flagellates) as well as various unicellular algae (chrysophytes) and muticellular algae (kelps and other brown algae).
Protists have traditionally been classified into the following non-monophyletic adaptive groups:
Discussion of Phylogenetic Relationships
General relationships among eukaryotes
Our understanding of the phylogenetic relationships among the eukaryotes is not yet resolved nor stable. Two large bodies of data have contributed most to our current understanding of the diversity and interrelationships of eukaryotic lineages.
Information derived from electron microscopy on the structure of the cells has revealed consistent patterns among groups for which the monophyly is not doubted (such as the ciliates or the red algae). The same approach has then been applied to many protists, and has revealed that there are about 80 patterns of organization (Patterson, 1999). These have now been clustered into about 60 lineages. Molecular analyses, when available, usually confirm these groupings.
The second body of data derives from comparative molecular data - initially focussing on the genes which code for small subunit ribosomal RNA. As it became evident that the insights might be distorted by problems in the methods of phylogenetic inference, and that our emerging insights (trees) were imprecise, so an increasing number of genes have been called upon to identify which elements of our understanding are secure and which are unrealiable. The position now is less confident than a decade earlier, and the means of resolving conflict among molecular insights has yet to be agreed upon (Philippe & Adoutte, 1998; Katz, 1999).
The most comprehensive molecular trees are still those based on analysis of 16S ribosomal RNA (e.g. Cavalier-Smith, 1993; Sogin & Silberman, 1998). The early molecular trees indicated that the earliest branches of eukaryote evolution are represented by microsporidia, trichomonads and diplomonads. These organisms lack dictyosomes, peroxisomes and conventional mitochondria. In addition, the organization of their cytoskeleton was simple and they had a relatively small number of membranous organelles when compared to more recently evolved taxa such as plants, animals and fungi. Organisms located higher in the tree had more organellar diversity, including the presence of dictyosomes, various membranous compartments and mitochondria and chloroplasts. The consistency of the molecular and structural insights led to models that these taxa were primitively amitochondriate, had derived early in eukaryote evolution, and could reveal to us the sequence in which the eukaryotic cell was assembled.
Early trees included an unresolved polytomy for the early-branching amitochondriate protists (Leipe et al., 1993). It was followed by the separation of the Euglenozoa (Euglena + other euglenids, trypanosomes + other kinetoplastids), a few other taxa such as the Heterolobosea (acrasid slime moulds, and the agents of amoebic meningitis - Naegleria), and a variety of amoeboid organisms (Sogin et al., 1996). The remaining organisms formed a cluster that was referred to as the eukaryotic crown and was interpreted as the nearly simultaneous separation of animals, plants, fungi and several complex protist assemblages (Knoll, 1992).
Within the last decade, it became increasingly evident that this understanding was not accurate (Roger, 1999). There is a problem that lineages which have shown rapid rates of evolution (have long branch lengths) are drawn together at the base of dendrograms created by programs that sought to interpret molecular data as evolutionary trees. Secondly, a variety of the 'amitochondriate' organisms have been shown to have genes for mitochondrial proteins suggesting that they are not primitively amitochondriate but secondarily amitochondriate. The presence of small membranous organelles in a number of these taxa suggests that they contain pre-mitochondria or reduced mitochondria. Enhanced molecular data provided evidence of different associations - the microsporidia were not organisms at the base of the tree but were a specialised kind of fungus and derived late in eukaryotic evolution, their structural simplicity being attributed to regression (Edlind et al., 1996; Keeling & Doolittle, 1996; Li et al., 1996; Germot et al., 1997; Edlind, 1998; Fast et al. 1999; Hirt et al., 1999; Keeling et al., 2000; Van de Peer et al., 2000). Molecular data also identified new candidates for the most primitive eukaryotes - Reclinomonas (an excavate flagellate) has a mitochondrial genome more replete with genes than any other and may be related to one of the first eukaryotes to acquire mitochondria (Lang et al., 1997). Yet molecular trees do not concur with each other (Katz, 1999). The consequence of these insights has been to demolish the model of the 1990's, but not to replace it with something better.
Yet, the intervening period has seen progress. At the beginning of the 1990's, we could recognise about 80 different types of eukaryotes (Patterson, 1994). In the intervening period, perhaps 10 further types of protists were being or have been added - either through the efforts of bioprospectors or through more detailed study of many of the underdescribed genera of protists. Yet only about 60 lineages are currently recognised. That is - about half of the lineages have found homes. We have now agreed that the sister group to the Metazoa are the collar-flagellates (choanoflagellates), and that these, together with the fungi and chytrids form a lineage (the opisthokonts), and that the opisthokonts contain two types of spore-forming organisms (Microsporidia and Myxozoa) that used to be considered as protists but are now seen as derived from multicellular organisms (Microsporidia from fungi, Myxozoa from coelenterates or bilateria); the ciliates, apicomplexan sporozoa and dinoflagellates are regarded as forming a lineage (the alveolates), the stramenopiles is now home to the brown algae, diatoms, chrysophytes, opalines, Blastocystis, some heliozoa, some heterotrophic flagellates and so on), and new groups - such as the excavates (including Giardia and the other diplomonads, retortamonads, and various heterotrophic flagellates such as Carpediemonas and the quadriflagellated Trimastix) - continue to be promoted. The resolution to the interrelationships of all eukaryotes looks as if it will reside in the piece by piece assembly of the jig-saw puzzle, rather than in the broad sweep approach which gave us so much confidence a decade ago.
Phylogenetic relationships among the opisthokonts
The term 'opisthokont' was introduced by Copeland (Copeland, 1956) for the chytrids - a small group of parasitic protists, now commonly included within the fungi. The name refers to the posterior (opistho) location of the flagellum (kont) in swimming cells. As comparative molecular biology indicated that the fungi and animals were related, so the term was applied to the (animals + fungi) clade (Cavalier-Smith & Chao, 1995). This is not entirely satisfactory, but an alternative name for the (animals + fungi) clade has yet to emerge.
The argument that the choanoflagellates gave rise to sponges and these in turn to the diploblastic and triploblastic animals is one with a long history, but an understanding of the origins of animals was impeded by spurious arguments based on reference to mythical ancestors (Hanson, 1977; Willmer, 1990). The relatedness of the collar flagellates (choanoflagellates) to the Metazoa was confirmed by comparative analyses of ribosomal RNA (Kim et al., 1999; Wainright et al., 1993), and the basal status of the sponges within Metazoa is widely accepted (Jenner & Schram 1999).
The relatedness of extended animal and fungal clades was not suggested by comparative morphology, but was revealed by comparative molecular biology (Baldauf & Palmer, 1993; Wainright et al., 1993; Sogin & Silberman, 1998; Baldauf, 1999). Subsequently, similarities in the anchorage systems of flagella of chytrid fungi and choanoflagellates have been identified, corroborating the molecular perspective (Moestrup, unpublished).
Microsporidian and myxosporan protists as members of the opisthokonts
Microsporidia are mostly unicellular intracellular parasites and have traditionally been classified within the protozoan group 'Sporozoa'. With the advent of molecular phylogeny, they were placed at the base of the tree of eukaryotes because of their gene structure and comparisons of small subunit ribosomal RNA. They also have a very simple cellular organization which corroborated this insight. The extension of comparative molecular biology to embrace more genes has led to the view that the microsporidia are a derived type of fungus, an argument supported by the presence of a distinctive signature sequence (Kamaishi et al., 1996; Keeling et al., 2000). There are no structural synapomorphies tying the microsporidia to the fungi or to a subset of the fungi.
Myxozoa (= Myxosporidia) were traditionally regarded as a type of protist which produces multicellular spores. The spore contains 'cells' which could eject filaments. In the 1970's it became evident that the appearance and development of the filaments co-incided with that of the nematocysts of the coelenterates (Metazoa). Molecular evidence has confirmed that Myxozoa are allied to the Metazoa, but molecular approaches are as yet unable to resolve particular relationships (Siddall et al., 1995; Smothers et al., 1994).
Symbioses, Endosymbioses and the Origin of the Eukaryotic State
Symbiosis is the process in which unrelated organisms come together and form a stable association - as do lichens (algae and fungi) and coral reefs (coelenterates and dinoflagellate algae). This idea was promulgated at the turn of the 20th century (Mereschkowsky, 1910) and promoted later in the same century by Lynn Margulis (Margulis, 1970). Symbiosis has been and still is an important driving force in the evolution of eukaryotes. Through this mechanism, complementary metabolic capabilities, life cycles, and competences of different organisms have been brough together to create an amalgam that is greater than the sum of the parts. Symbiosis has provided a selective advantage to organisms when responding to changing environments. The close proximity of partners in symbiosis creates opportunities for coevolution of genomes and for the lateral transfer of genetic information between compartments or between evolutionary lineages. Microorganisms that engage in symbioses may be either extracellular or intracellular. Examples of extracellular symbionts include lichenized fungi and various bacteria which reside within metazoan digestive systems. There are also extracellular bacteria which live on the surface of eukaryotic microorganisms.

Symbiotic bacteria (one is indicated by the arrow) coat the surface of a devescovinid flagellate (a member of the parabasalids), its flagella are to the right.
Photograph copyright © 2000 Mitchell L. Sogin
Intracellular symbionts are referred to as endosymbionts, and their impact upon eukaryote evolution has been especially profound. Endosymbiosis was important in the evolution of even the earliest diverging eukaryotic cells. In the most radical views of eukaryotic origins it is suggested (but not proven) that formation of the eukaryotic cell was a consequence of genome fusions between a host cell and endosymbionts representing distinct evolutionary lineages (Sogin, 1991; Forterre et al., 1992; Lake & Rivera, 1994; Golding & Gupta, 1995; Katz, 1998, 1999).
The role of endosymbionts in the origins of mitochondria and chloroplasts is more convincing. The ancestors of these organelles were initially independent bacterial organisms which were acquired by or invaded early eukaryotes. It is believed that by enriching the array of metabolic processes available to the cell, the symbionts conferred a selective advantage to ancestral eukaryotes. The consortium was favoured, and the endosymbionts became integrated in the operations of the cell as organelles.
In the case of mitochondria, ultrastructure and molecular data indicate that they are derived from the alpha-proteo bacteria (Yang et al., 1985). This particular association provided a means for early eukaryotic cells--previously limited to anaerobic metabolism--to carry out aerobic respiration. The benefit is that this association allowed oxygen to be used as a terminal electron acceptor, and the energy derived from ingested food increased by a factor of almost 20. There is now considerable controversy about the timing of this endosymbiotic event and whether there are any remnants of pre-mitochondrial euklaryotes. Ribosomal RNA trees indicate that the earliest diverging eukaryotes (e.g. diplomonads, trichomonads) lack mitochondria and this was thought to indicate that these taxa were primitively without mitochondria (see Discussion of Phylogenetic Relationships). However, there is now increasing evidence that genes normally found in alpha-proteo bacteria are also present in the nucleus of amitochondriate prostists and that mitochondria were present much earlier than previously thought, but have now been lost from some early eukaryotes (Roger, 1999).
The origins of chloroplasts are also rooted in endosymbiotic processes. Probably later than the acquisition of mitochondria, a cyanobacterium took up residence in an ancient eukaryote. These primary endosymbionts were destined to become the chloroplasts which are found in eukaryotic algae. Their role may have been to add autotrophic competency to heterotrophic cells, or perhaps to provide oxygen to improve efficiency of metabolism. Chloroplasts have proven to be promiscuous, and some eukaryotic algae acquired their chloroplasts through secondary endosymbioses in which the plastid of one eukaryotic algal species was transferred to and became established in different eukaryotic clades (Delwiche, 1999). At this time, there is no evidence that any of the other familiar components of eukaryotic cells were acquired by symbiosis.
References
Arisue, N., M. Hasegawa, and T. Hashimoto. 2005. Root of the Eukaryota tree as inferred from combined maximum likelihood analyses of multiple molecular sequence data. Molecular Biology and Evolution 22(3):409-420.
Baldauf, S. L. 1999. A search for the origins of animals and fungi: Comparing and combining molecular data. American Naturalist 154(suppl.):S178-S188.
Baldauf, S. L. and W. F. Doolittle. 1997. Origin and evolution of the slime molds (Mycetozoa). Proceedings of the National Academy of Sciences (USA) 94:12007-12012.
Baldauf, S. L. and J. D. Palmer. 1993. Animals and fungi are each other's closest relatives: Congruent evidence from multiple proteins. Proceedings of the National Academy of Sciences (USA) 90:11558-11562.
Borchiellini C., N. Boury-Esnault, J. Vacelet, and Y. Le Parco. 1998. Phylogenetic analysis of the Hsp70 sequences reveals the monophyly of metazoa and specific phylogenetic relationships between animals and fungi. Molecular Biology and Evolution 15:647-655.
Budin, K. and H. Philippe. 1998. New insights into the phylogeny of eukaryotes based on Ciliate Hsp70 sequences. Molecular Biology and Evolution 15:943-956.
Canning, E. U. 1998. Evolutionary relationships of Microsporidia. Pages 77-90 in Evolutionary Relationships among Protozoa (G. H. Coombs, K. Vickerman, M .A. Sleigh, and A. Warren, eds.) Chapman & Hall, London.
Cavalier-Smith, T. 1993. Kingdom Protozoa and its 18 phyla. Microbiol. Rev. 57:953-94.
Cavalier-Smith, T. and Chao, E. E. 1995. The opalozoan Apusomonas is related to the common ancestor of animals, fungi and choanoflagellates. Proceedings of the Royal Society of London Series B 261:1-6.
Clark C. G. and A. J. Roger. 1995. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proceedings of the National Academy of Sciences (USA) 92:6518-6521.
Copeland, H. F. 1956. The Classification of Lower Organisms. Pacific Books, Palo Alto, California.
Delwiche, C. F. 1999. Tracing the thread of plastid diversity through the tapistry of life. American Naturalist 154 (suppl.):S164-S177.
Douzery, E. J. P., E. A. Snell, E. Bapteste, F. Delsuc, and H. Philippe. 2004. The timing of eukaryotic evolution: Does a relaxed molecular clock reconcile proteins and fossils? Proceedings of the National Academy of Sciences (USA) 101(43):15386-15391.
Edlind, T. D. 1998. Phylogenetics of protozoan tubulin with reference to the amitochondriate eukaryotes. Pages 91-108 in Evolutionary Relationships Among Protozoa (G. H. Coombs, K. Vickerman, M. A. Sleigh, and A. Warren, eds.) Chapman & Hall, London.
Edlind, T. D., J. Li, G. S. Visvesvara, M. H. Vodkin, G. L. McLaughlin, and S. K. Katiyar. 1996. Phylogenetic analysis of beta-tubulin sequences from amitochondrial protozoa. Molecular Phylogenetics and Evolution 5:359-367.
Embley T. M. and R. P. Hirt. 1998. Early branching eukaryotes? Curr. Opinion Gen. Dev. 8:624-629.
Fast, N. M., J. M. Logsdon, and W. F. Doolittle. 1999. Phylogenetic analysis of the TATA box binding protein (TBP) gene from Nosema locustae: evidence for a microsporidia-fungi relationship and spliceosomal intron loss. Molecular Biology and Evolution 16:1415-1419.
Forterre, P., N. Benachenhou-Lahfa, F. Confalonieri, M. Duguet, C. Elie and B. Labedan. 1992. The nature of the last universal ancestor and the root of the tree of life, still open questions. Biosystems 28:15-32.
Germot, A., H. Philippe, and H. Le Guyader. 1997. Evidence for loss of mitochondria in Microsporidia from a mitochondrial-type HSP70 in Nosema locustae. Molecular and Biochemical Parasitology 87:159-168.
Golding, G. B. and R. S. Gupta. 1995. Protein-based phylogenies support a chimeric origin for the eukaryotic genome. Molecular Biology and Evolution 12:1-6.
Hampl, V., D. S. Horner, P. Dyal, J. Kulda, J. Flegr, P. G. Foster, and T. M. Embley. 2005. Inference of the phylogenetic position of oxymonads based on nine genes: Support for Metamonada and Excavata. Molecular Biology and Evolution 22(12):2508-2518.
Hanson, E. D. 1977. The Origin and Early Evolution of Animals. Wesleyan University Press, Middletown, Conn.
Hashimoto, T., Y. Nakamura, T. Kamaishi, and M. Hasegawa. 1997. Early evolution of eukaryotes inferred from protein phylogenies of translation elongation factors 1 alpha and 2. Archiv f?r Protistenkunde 148:287-295.
Hirt, R. P., B. Healy, C. R. Vossbrinck, E. U. Canning, and T. M. Embley. 1997. A mitochondrial Hsp70 orthologue in Vairimorpha necatrix: Molecular evidence that microsporidia once contained mitochondria. Current Biology 7:995-998.
Hirt, R. P. and D. Horner (eds.) 2004. Organelles, Genomes and Eukaryote Evolution. Taylor & Francis, London.
Hirt, R. P., J. M. Logsdon, Jr., B. Healy, M. W. Dorey, W. F. Doolittle, and T. M. Embley. 1999. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proceedings of the National Academy of Sciences (USA) 96:580-585.
Huang, J., Y. Xu, and J. P. Gogarten. 2005. The presence of a haloarchaeal type tyrosyl-tRNA synthetase marks the opisthokonts as monophyletic. Molecular Biology and Evolution 22:2142-2146.
Jenner, R. A. and F. R. Schram. 1999. The grand game of metazoan phylogeny: rules and strategies. Biological Reviews 74:121-142.
Kamaishi, T., T. Hashimoto, Y. Nakamura, F. Nakamura, S. Murata, N. Okada, K. Okamoto, and M. Hasegawa. 1996. Protein phylogeny of translation elongation factor EF-1alpha suggests microsporidians are extremely ancient eukaryotes. Journal of Molecular Evolution 42:257-263.
Katz, L. A. 1998. Changing perspectives on the origin of eukaryotes. Trends Ecol. Evol. 13:493-497.
Katz, L. A. 1999. The tangled web: gene genealogies and the origin of eukaryotes. American Naturalist 154 (suppl.):S137-S145.
Keeling, P. J. 1998. A kingdom's progress: Archezoa and the origin of eukaryotes. BioEssays 20:87-95.
Keeling, P. J. 2004. Diversity and evolutionary history of plastids and their hosts. American Journal of Botany 91:1481-1493.
Keeling, P. J., G. Burger, D. G. Durnford, B. F. Lang, R. W. Lee, R. E. Pearlman, A. J. Roger and M. W. Gray. 2005. The tree of eukaryotes. Trends in Ecology & Evolution 20(12):670-676.
Keeling, P. J. and W. F. Doolittle. 1996. Alpha-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Molecular Biology and Evolution 13:1297-1305.
Keeling, P. J., M. A. Luker, and J. D. Palmer. 2000. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Molecular Biology and Evolution 17:23-31.
Keeling, P. J. and G. I. McFadden. 1998. Origins of microsporidia. Trends Microbiol. 6:19-23.
Keeling, P. J. and J. D. Palmer. 2000. Phylogeny - Parabasalian flagellates are ancient eukaryotes. Nature 405:635-637.
Kim, J., W. Kim, and C. W. Cunningham. 1999. A new perspective on lower metazoan relationships from 18S rDNA sequences. Molecular Biology and Evolution 16:423-427.
Knoll, A. H. 1992. The early evolution of eukaryotes: a geological perspective. Science 256:622-627.
Kumar, S. and A. Rzhetsky. 1996. Evolutionary relationships of eukaryotic kingdoms. Journal of Molecular Evolution 42:183-193.
Lake, J. A. and M. C. Rivera. 1994. Was the nucleus the first endosymbiont? Proceedings of the National Academy of Sciences (USA) 91:2880-2881.
Lang, B. F., G. Burger, C. J. O'Kelly, R. Cedergren, G. B. Golding, C. Lemieux, D. Sankoff, M. Turmel, and M. W. Gray. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387:493-497.
Lang, B. F., M. W. Gray, and G. Burger. 1999. Mitochondrial genome evolution and the origin of eukaryotes. Annual Review of Genetics 33:351-397.
Leipe, D., J. H. Gunderson, T. A. Nerad and M. L. Sogin. 1993. Small subunit ribosomal RNA of Hexamita inflata and the quest for the first branch in the eukaryotic tree. Mol. Biochem. Parasitol. 59:41-48.
Li, J., S. K. Katiyar, A. Hamelin, G. S. Visvesvara, and T. D. Edlind. 1996. Tubulin genes from AIDS-associated microsporidia and implications for phylogeny and benzimidazole sensitivity. Molecular and Biochemical Parasitology 78:289-295.
Lipscomb D. L., J. S. Farris, M. Kallersjo and A. Tehler. 1998. Support, ribosomal sequences and the phylogeny of the eukaryotes. Cladistics 14:303-338.
Maldonado, M. 2004. Choanoflagellates, choanocytes, and animal multicellularity. Invertebrate Biology 123:1?22.
Margulis, L. 1970. Origin of Eukaryotic Cells. Yale University Press.
Margulis, L., J. O. Corliss, M. Melkonian and D. J. Chapman. 1990. Handbook of Protoctista. Jones and Bartlett Publishers, Boston.
Mereschkowsky, C. 1910. Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis. Eine neue Lehre von der Entstehung der Organismen. Biologisches Centralblatt 30: 278-303, 321-347, 353-367.
Moreira, D., H. Le Guyader, and H. Philippe. 2000. The origin of red algae and the evolution of chloroplasts. Nature 405:69-72.
Morin, L. 2000. Long branch attraction effects and the status of "basal eukaryotes": Phylogeny and structural analysis of the ribosomal RNA gene cluster of the free-living diplomonad Trepomonas agilis. Journal of Eukaryotic Microbiology 47:167-177.
Morris, P. J. 1993. The developmental role of the extracellular matrix suggests a monophyletic origin of the Kingdom Animalia. Evolution 47:152-165.
Narbonne, G. M. 2004. Modular construction of early Ediacaran complex life forms. Science 305(5687):1141-1144.
Nozaki, H., M. Matsuzaki, M. Takahara, O. Misumi, H. Kuroiwa, M. Hasegawa, T. Shin-i, Y. Kohara, N. Ogasawara, and T. Kuroiwa. 2003. The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids. Journal of Molecular Evolution 56(4):485-497.
Patterson, D. J. 1994. Protozoa: Evolution and Systematics. Pages 1-14 in Progress in Protozoology. Proceedings of the IX International Congress of Protozoology, Berlin 1993. (K. Hausmann and N. H?lsmann, eds.) Gustav Fischer Verlag, Stuttgart, Jena, New York.
Patterson, D. J. 1999. The diversity of eukaryotes. American Naturalist 154 (suppl.):S96-S124.
Patterson, D. J. and M. L. Sogin. 1992. Eukaryote origins and protistan diversity. Pages 13-46 in The Origin and Evolution of Prokaryotic and Eukaryotic Cells (H. Hartman and K. Matsuno, eds.) World Scientific Pub. Co. NJ.
Philippe, H. and A. Adoutte. 1998. The molecular phylogeny of Eukaryota: solid facts and uncertainties. Pages 25-56 in Evolutionary Relationships among Protozoa (G. H. Coombs, K. Vickerman, M. A. Sleigh, and A. Warren, eds.) Chapman & Hall, London.
Philippe, H. and A. Germot. 2000. Phylogeny of eukaryotes based on ribosomal RNA: Long-branch attraction and models of sequence evolution. Molecular Biology and Evolution 17:830-834.
Philippe, H., P. Lopez, H. Brinkmann, K. Budin, A. Germot, J. Laurent, D. Moreira, M. Muller, and H. Le Guyader. 2000. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proceedings of the Royal Society of London Series B 267:1213-1221.
Philippe, H., E. A. Snell, E. Bapteste, P. Lopez, P. W. H. Holland, and D. Casane. 2004. Phylogenomics of eukaryotes: impact of missing data on large alignments. Molecular Biology and Evolution 21(9):1740-1752.
Ragan, M. A. and R. R. Gutell. 1995. Are red algae plants? Botanical Journal of the Linnean Society 118:81-105.
Ribeiro, S. and G. B. Golding. 1998. The mosaic nature of the eukaryotic nucleus. Molecular Biology and Evolution 15:779-788.
Roger, A. J. 1999. Reconstructing early events in eukaryotic evolution. American Naturalist 154 (suppl.):S146-S163.
Roger, A. J., O. Sandblom, W. F. Doolittle, and H. Philippe. 1999. An evaluation of elongation factor 1 alpha as a phylogenetic marker for eukaryotes. Molecular Biology and Evolution 16:218-233.
Schlegel, M. 2003. Phylogeny of Eukaryotes recovered with molecular data: highlights and pitfalls. European Journal of Protistology 39:113-122.
Sch?tze J., A. Krasko, M. R. Custodio, S. M. Efremova, I. M. M?ller and W. E. G. M?ller. 1999. Evolutionary relationships of Metazoa within the eukaryotes based on molecular data from Porifera. Proceedings of the Royal Society of London Series B 266:63-73.
Siddall, M. E., D. S. Martin, D. Bridge, S. S. Desser, and D.K. Cone. 1995. The demise of a phylum of protists: phylogeny of Myxozoa and other parasitic Cnidaria. J. Parasitol. 81:961-967.
Smothers, J. F., C. D. van Dohlen, L. H. Smith, and R. D. Spall. 1994. Molecular evidence that the myxozoan protists are metazoans. Science 265:1719-1721.
Sogin, M. L. 1991. Early evolution and the origin of eukaryotes. Current Opinion in Genetics and Development 1:457-463.
Sogin, M. L., H. G. Morrison, G. Hinkle and J. D. Silberman. 1996. Ancestral relationships of the major eukaryotic lineages. Microbiologia SEM 12:17-28.
Sogin, M. L. and J. D. Silberman. 1998. Evolution of the protists and protistan parasites from the perspective of molecular systematics. International Journal of Parasitology 28:11-20.
Stechmann, A. and T. Cavalier-Smith. 2002. Rooting the eukaryote tree by using a derived gene fusion. Science 297:89-91.
Steenkamp, E. T., J. Wright, and S. L. Baldauf. 2006. The protistan origins of animals and fungi. Molecular Biology and Evolution 23:93-106.
Stiller, J. W., E. C. S. Duffield and B. D. Hall. 1998. Amitochondriate amoebae and the evolution of DNA-dependent RNA polymerase II. Proceedings of the National Academy of Sciences (USA) 95:11769-11774.
Stiller, J. W. and B. D. Hall. 1997. The origin of red algae: Implications for plastid evolution. Proceedings of the National Academy of Sciences (USA) 94:4520-4525.
Stiller, J. W. and B. D. Hall. 1999. Long-branch attraction and the rDNA model of early eukaryotic evolution. Molecular Biology and Evolution 16:1270-1279.
Stiller, J. W, J. Riley, and B. D. Hall. 2001. Are red algae plants? A critical evaluation of three key molecular data sets. Journal of Molecular Evolution 52(6):527-539.
Taylor F. J. R. 1999. Ultrastructure as a control for protistan molecular phylogeny. American Naturalist 154(suppl.):S125-S136.
Van de Peer, Y., A. Ben Ali, and A. Meyer. 2000. Microsporidia: accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene 246:1-8.
Van de Peer, Y. and R. de Wachter. 1997. Evolutionary relationships among the eukaryotic crown taxa taking into account site-to-site rate variation in 18S rRNA. Journal of Molecular Evolution 45:619-630.
Van de Peer, Y., G. Van der Auwera and R. DeWachter. 1996. The evolution of stramenopiles and alveolates as derived by 'substitution rate calibration' of small ribosomal subunit RNA. Journal of Molecular Evolution 42:201-210.
Vellai T. and G. Vida. 1999. The origin of eukaryotes: the difference between prokaryotic and eukaryotic cells. Proceedings of the Royal Society of London Series B 266:1571-1577.
Wainright, P. O., G. Hinkle, M. L. Sogin, and S. K. Stickel. 1993. Monophyletic origin of the Metazoa: an evolutionary link with fungi. Science 260:340-342.
Wainright, P. O., D. J. Patterson, and M. L. Sogin. 1994. Monophyletic origin of animals: a shared ancestry with fungi. Pages 39-53 in Molecular Evolution of Physiological Processes. Society of General Physiologists Series No. 49. (D. M. Farmborough, ed.) Rockefeller Press, New York.
Willmer, P. 1990. Invertebrate Relationships: Patterns in Animal Evolution. Cambridge University Press, Cambridge, UK.
Yang, D., Y. Oyaizu, H. Oyaizu, G. J. Olsen, and C. R. Woese. 1985. Mitochondrial origins. Proceedings of the National Academy of Sciences (USA) 82:4443-4447.
Information on the Internet
- Eu-Tree. Assembling the Tree of Eukaryotic Diversity.
- Protsville. Protist Research Laboratory, University of Sydney, Australia.
- Protist Information Server. Japan Science and Technology Corporation.
- Eukaryota: Systematics. Museum of Paleontology, University of California, Berkeley, USA.
- Exploring Early Eukaryotic Evolution: Diversity and Relationships Among Novel Deep-Branching Lineages . Virginia Edgcomb, Andrew Roger, Alastair G.B. Simpson, Jeffrey Silberman and Mitchell Sogin, Marine Biological Laboratory, Woods Hole, USA.
- Eukaryotes in extreme environments. Dave Roberts, the Natural History Museum, London, UK.
- Protist Image Data. Molecular Evolution and Organelle Genomics program at the University of Montreal, Canada.
Title Illustrations
| Scientific Name | Cereus giganteus |
|---|---|
| Location | Tucson, Arizona, U.S.A. |
| Comments | Saguaros, the giant columnar cacti of the Sonoran desert of northwestern Mexico and the southwestern U.S.A. |
| Specimen Condition | Live Specimen |
| Life Cycle Stage | adults |
| Copyright | © 1994 |
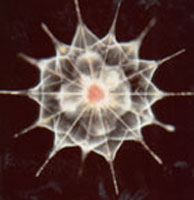
| Scientific Name | Acantharea |
|---|---|
| Comments | A star-shaped protist that lives in marine habitats - its yellow colour comes from symbiotic algae living inside it. |
| Copyright | © 1998 Linda Amaral Zettler |
| Scientific Name | Ithomiini, Senecio |
|---|---|
| Location | Monteverde, Costa Rica |
| Comments | Ithomiine butterfly feeding on Senecio flowers |
| Specimen Condition | Live Specimen |
| Copyright | © 1996 Greg and Marybeth Dimijian |
About This Page
David J. Patterson
The Josephine Bay Paul Center in Comparative Molecular Biology and Evolution
Marine Biological Laboratory
7 MBL Street
Woods Hole, Massachusetts 02543
USA
Mitchell L. Sogin
The Josephine Bay Paul Center in Comparative Molecular Biology and Evolution
Marine Biological Laboratory
7 MBL Street
Woods Hole, Massachusetts 02543
USA
Correspondence regarding this page should be directed to David J. Patterson at
Page copyright © 2000 David J. Patterson and Mitchell L. Sogin
- First online 08 September 2000
Citing this page:
Patterson, David J. and Sogin, Mitchell L. 2000. Eukaryotes. Eukaryota, Organisms with nucleated cells. Version 08 September 2000 (complete). http://tolweb.org/Eukaryotes/3/2000.09.08 in The Tree of Life Web Project, http://tolweb.org/